Publications
Publications
A list of my academic publications. Selected works are highlighted.
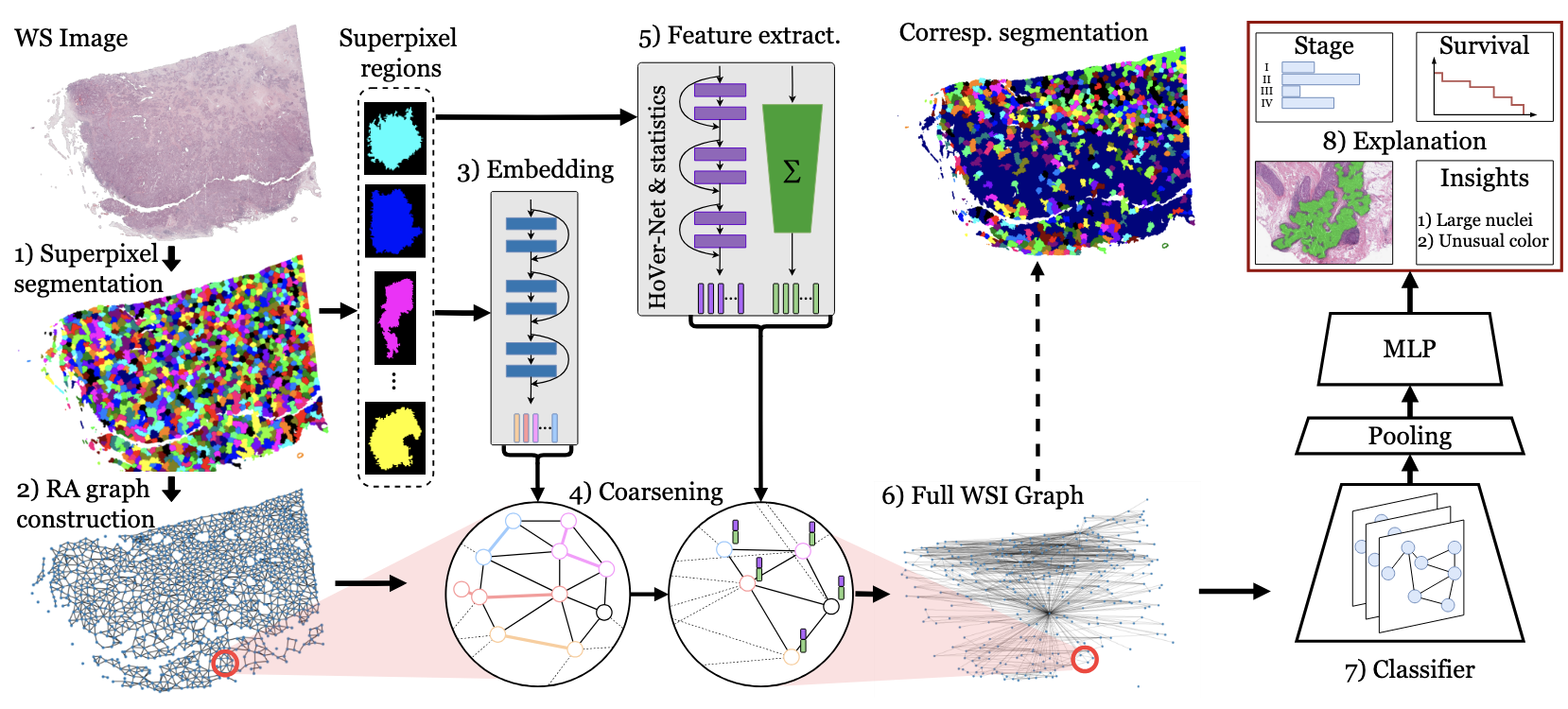
A Graph-Based Framework for Interpretable Whole Slide Image Analysis
Alexander Weers, Alexander Berger, Laurin Lux, Peter Schüffler, Daniel Rueckert, Johannes Paetzold
arXiv preprint arXiv:2503.11846, 2025
Abstract
The histopathological analysis of whole-slide images (WSIs) is fundamental to cancer diagnosis but is a time-consuming and expert-driven process. While deep learning methods show promising results, dominant patch-based methods artificially fragment tissue, ignore biological boundaries, and produce black-box predictions. We overcome these limitations with a novel framework that transforms gigapixel WSIs into biologically-informed graph representations and is interpretable by design. Our approach builds graph nodes from tissue regions that respect natural structures, not arbitrary grids. We introduce an adaptive graph coarsening technique, guided by learned embeddings, to efficiently merge homogeneous regions while preserving diagnostically critical details in heterogeneous areas. Each node is enriched with a compact, interpretable feature set capturing clinically-motivated priors. A graph attention network then performs diagnosis on this compact representation. We demonstrate strong performance on challenging cancer staging and survival prediction tasks. Crucially, our resource-efficient model (>13x fewer parameters and >300x less data) achieves results competitive with a massive foundation model, while offering full interpretability through feature attribution.
View paper →
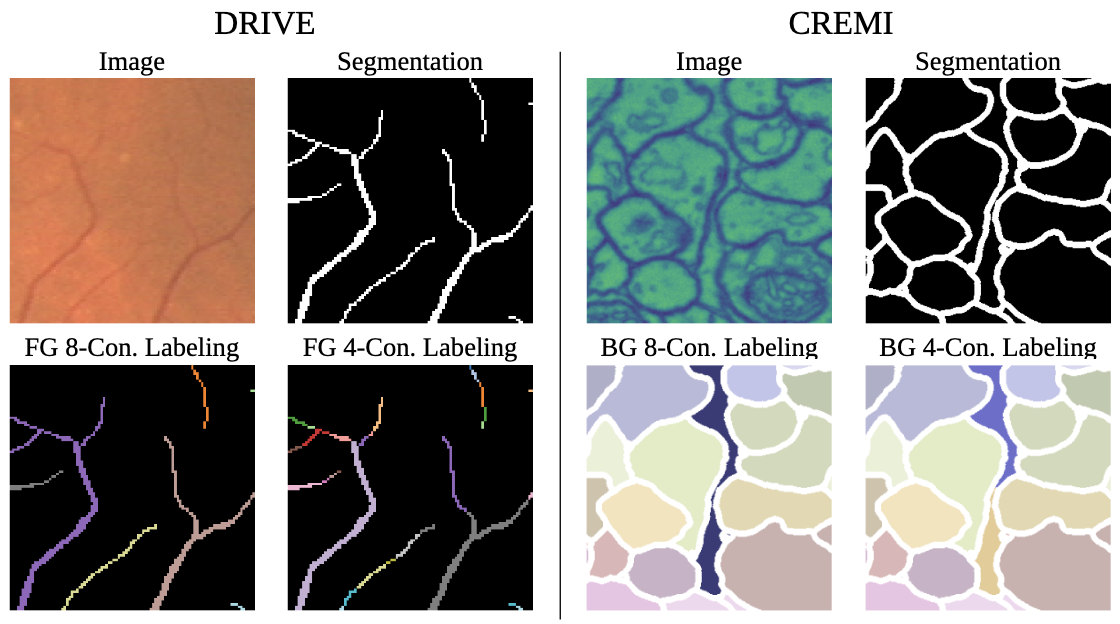
Pitfalls of topology-aware image segmentation
Alexander Berger, Laurin Lux, Alexander Weers, Martin Menten, Daniel Rueckert, Johannes Paetzold
International Conference on Information Processing in Medical Imaging, 2025
Abstract
Topological correctness, i.e., the preservation of structural integrity and specific characteristics of shape, is a fundamental requirement for medical imaging tasks, such as neuron or vessel segmentation. Despite the recent surge in topology-aware methods addressing this challenge, their real-world applicability is hindered by flawed benchmarking practices. In this paper, we identify critical pitfalls in model evaluation that include inadequate connectivity choices, overlooked topological artifacts in ground truth annotations, and inappropriate use of evaluation metrics. Through detailed empirical analysis, we uncover these issues profound impact on the evaluation and ranking of segmentation methods. Drawing from our findings, we propose a set of actionable recommendations to establish fair and robust evaluation standards for topology-aware medical image segmentation methods.
View paper →
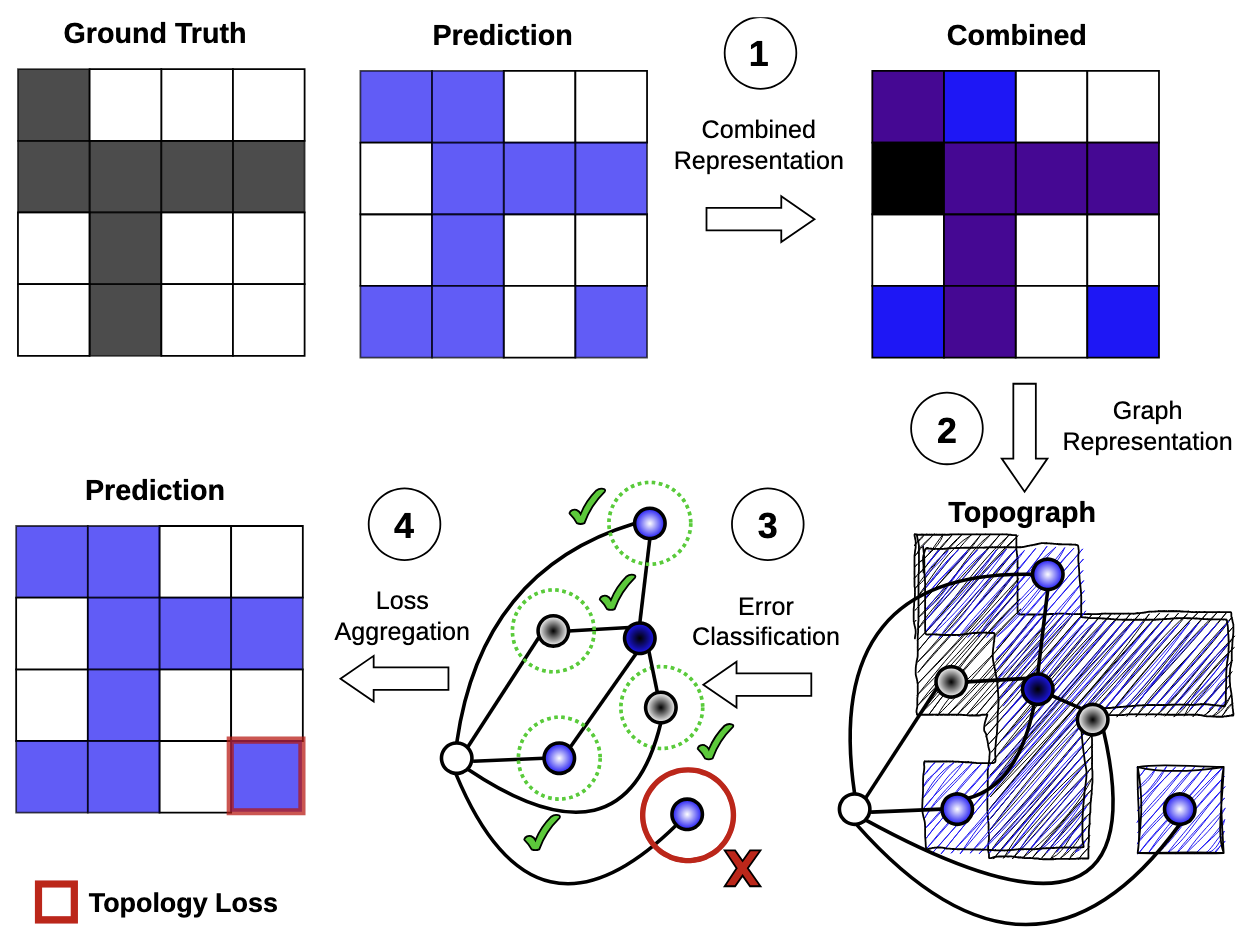
Topograph: An efficient graph-based framework for strictly topology preserving image segmentation
Laurin Lux, Alexander Berger, Alexander Weers, Nico Stucki, Daniel Rueckert, Ulrich Bauer, Johannes Paetzold
ICLR, 2025
Abstract
Topological correctness plays a critical role in many image segmentation tasks, yet most networks are trained using pixel-wise loss functions, such as Dice, neglecting topological accuracy. Existing topology-aware methods often lack robust topological guarantees, are limited to specific use cases, or impose high computational costs. In this work, we propose a novel, graph-based framework for topologically accurate image segmentation that is both computationally efficient and generally applicable. Our method constructs a component graph that fully encodes the topological information of both the prediction and ground truth, allowing us to efficiently identify topologically critical regions and aggregate a loss based on local neighborhood information. Furthermore, we introduce a strict topological metric capturing the homotopy equivalence between the union and intersection of prediction-label pairs. We formally prove the topological guarantees of our approach and empirically validate its effectiveness on binary and multi-class datasets. Our loss demonstrates state-of-the-art performance with up to fivefold faster loss computation compared to persistent homology methods.
View paper →

